METAGENE-1
Model Overview
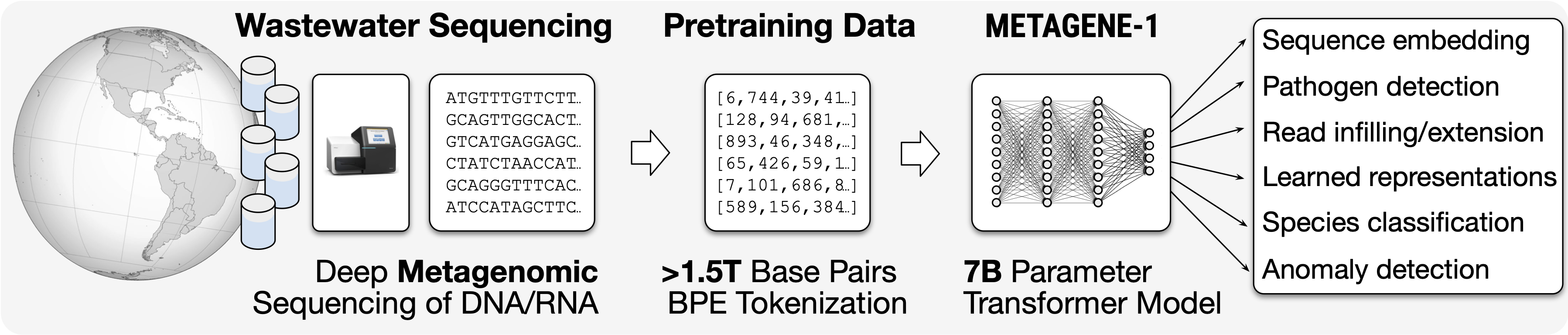
METAGENE-1 is a 7B parameter metagenomic foundation model designed for pandemic monitoring, trained on over 1.5T base pairs of DNA and RNA sequenced from wastewater.
METAGENE-1 is a 7-billion-parameter autoregressive transformer language model, which we refer to as a metagenomic foundation model, that was trained on a novel corpus of diverse metagenomic DNA and RNA sequences comprising over 1.5 trillion base pairs. This dataset is sourced from a large collection of human wastewater samples, processed and sequenced using deep metagenomic (next-generation) sequencing methods. Unlike genomic models that focus on individual genomes or curated sets of specific species, the aim of METAGENE-1 is to capture the full distribution of genomic information present across the human microbiome. After pretraining, this model is designed to aid in tasks in the areas of biosurveillance, pandemic monitoring, and pathogen detection.
We carry out byte-pair encoding (BPE) tokenization on our dataset, tailored for metagenomic sequences, and then pretrain our model. We detail the pretraining data, tokenization strategy, and model architecture, highlighting the considerations and design choices that enable the effective modeling of metagenomic data, in our technical report.
Usage
import torch
from transformers import AutoTokenizer, AutoModelForCausalLM
# Load the tokenizer and model
tokenizer = AutoTokenizer.from_pretrained("metagene-ai/METAGENE-1")
model = AutoModelForCausalLM.from_pretrained("metagene-ai/METAGENE-1", torch_dtype=torch.bfloat16)
# Example input sequence
input_sequence = "TCACCGTTCTACAATCCCAAGCTGGAGTCAAGCTCAACAGGGTCTTC"
# Tokenize the input sequence and remove the [EOS] token for generation
input_tokens = tokenizer.encode(input_sequence, return_tensors="pt", add_special_tokens=False)
# Generate output from the model
generated_tokens = model.generate(input_tokens, max_length=32)
# Decode the generated output and clean up the result
generated_sequence = tokenizer.decode(generated_tokens[0], skip_special_tokens=True)
generated_sequence = generated_sequence.replace(" ", "").replace("_", "")
# Generated output: A Hexamita inflata 5.8S ribosomal RNA gene sequence
print(f"🔬 Generated Sequence:\n{generated_sequence}")
# TCACCGTTCTACAATCCCAAGCTGGAGTCAAGCTCAACAGGGTCTTCTTGCCCCGCTGAGGGTTACACTCGCCCGTTCCCGAGTCTGTGGTTTCGCGAAGATATGACCAGGGACAGTAAGAACC
Benchmark Performance
We evaluate METAGENE-1 across three tasks: pathogen detection, zero-shot embedding benchmarks (Gene-MTEB), and genome understanding (GUE), achieving state-of-the-art performance on most benchmarks. For more details, check out our paper.
Pathogen Detection
The pathogen detection benchmark evaluates METAGENE-1’s ability to classify sequencing reads as human pathogens or non-pathogens across four distinct datasets, each derived from different sequencing deliveries and designed to mimic real-world conditions with limited training data.
| DNABERT-2 | DNABERT-S | NT-2.5b-Multi | NT-2.5b-1000g | METAGENE-1 | |
|---|---|---|---|---|---|
| Pathogen-Detect (avg.) | 87.92 | 87.02 | 82.43 | 79.02 | 92.96 |
| Pathogen-Detect-1 | 86.73 | 85.43 | 83.80 | 77.52 | 92.14 |
| Pathogen-Detect-2 | 86.90 | 85.23 | 83.53 | 80.38 | 90.91 |
| Pathogen-Detect-3 | 88.30 | 89.01 | 82.48 | 79.83 | 93.70 |
| Pathogen-Detect-4 | 89.77 | 88.41 | 79.91 | 78.37 | 95.10 |
Gene-MTEB
The Gene-MTEB benchmark evaluates METAGENE-1’s ability to produce high-quality, zero-shot genomic representations through eight classification and eight clustering tasks.
| DNABERT-2 | DNABERT-S | NT-2.5b-Multi | NT-2.5b-1000g | METAGENE-1 | |
|---|---|---|---|---|---|
| Human-Virus (avg.) | 0.564 | 0.570 | 0.675 | 0.710 | 0.775 |
| Human-Virus-1 | 0.594 | 0.605 | 0.671 | 0.721 | 0.828 |
| Human-Virus-2 | 0.507 | 0.510 | 0.652 | 0.624 | 0.742 |
| Human-Virus-3 | 0.606 | 0.612 | 0.758 | 0.740 | 0.835 |
| Human-Virus-4 | 0.550 | 0.551 | 0.620 | 0.755 | 0.697 |
| HMPD (avg.) | 0.397 | 0.403 | 0.449 | 0.451 | 0.465 |
| HMPD-single | 0.292 | 0.293 | 0.285 | 0.292 | 0.297 |
| HMPD-disease | 0.480 | 0.486 | 0.498 | 0.489 | 0.542 |
| HMPD-sex | 0.366 | 0.367 | 0.487 | 0.476 | 0.495 |
| HMPD-source | 0.451 | 0.465 | 0.523 | 0.545 | 0.526 |
| HVR (avg.) | 0.479 | 0.479 | 0.546 | 0.524 | 0.550 |
| HVR-p2p | 0.548 | 0.550 | 0.559 | 0.650 | 0.466 |
| HVR-s2s-align | 0.243 | 0.241 | 0.266 | 0.293 | 0.267 |
| HVR-s2s-small | 0.373 | 0.372 | 0.357 | 0.371 | 0.467 |
| HVR-s2s-tiny | 0.753 | 0.753 | 1.000 | 0.782 | 1.000 |
| HMPR (avg.) | 0.347 | 0.351 | 0.348 | 0.403 | 0.476 |
| HMPR-p2p | 0.566 | 0.580 | 0.471 | 0.543 | 0.479 |
| HMPR-s2s-align | 0.127 | 0.129 | 0.144 | 0.219 | 0.140 |
| HMPR-s2s-small | 0.419 | 0.421 | 0.443 | 0.459 | 0.432 |
| HMPR-s2s-tiny | 0.274 | 0.274 | 0.332 | 0.391 | 0.855 |
| Global Average | 0.475 | 0.479 | 0.525 | 0.545 | 0.590 |
GUE
Next, we evaluate METAGENE-1 on the GUE multi-species classification benchmark proposed in DNABERT-2. This experiment is designed to assess the viability of METAGENE-1 as a general-purpose genome foundation model.
| CNN | HyenaDNA | DNABERT | NT-2.5B-Multi | DNABERT-2 | METAGENE-1 | |
|---|---|---|---|---|---|---|
| TF-Mouse (avg.) | 45.3 | 51.0 | 57.7 | 67.0 | 68.0 | 71.4 |
| 0 | 31.1 | 35.6 | 42.3 | 63.3 | 56.8 | 61.5 |
| 1 | 59.7 | 80.5 | 79.1 | 83.8 | 84.8 | 83.7 |
| 2 | 63.2 | 65.3 | 69.9 | 71.5 | 79.3 | 83.0 |
| 3 | 45.5 | 54.2 | 55.4 | 69.4 | 66.5 | 82.2 |
| 4 | 27.2 | 19.2 | 42.0 | 47.1 | 52.7 | 46.6 |
| TF-Human (avg.) | 50.7 | 56.0 | 64.4 | 62.6 | 70.1 | 68.3 |
| 0 | 54.0 | 62.3 | 68.0 | 66.6 | 72.0 | 68.9 |
| 1 | 63.2 | 67.9 | 70.9 | 66.6 | 76.1 | 70.8 |
| 2 | 45.2 | 46.9 | 60.5 | 58.7 | 66.5 | 65.9 |
| 3 | 29.8 | 41.8 | 53.0 | 51.7 | 58.5 | 58.1 |
| 4 | 61.5 | 61.2 | 69.8 | 69.3 | 77.4 | 77.9 |
| EMP (avg.) | 37.6 | 44.9 | 49.5 | 58.1 | 56.0 | 66.0 |
| H3 | 61.5 | 67.2 | 74.2 | 78.8 | 78.3 | 80.2 |
| H3K14ac | 29.7 | 32.0 | 42.1 | 56.2 | 52.6 | 64.9 |
| H3K36me3 | 38.6 | 48.3 | 48.5 | 62.0 | 56.9 | 66.7 |
| H3K4me1 | 26.1 | 35.8 | 43.0 | 55.3 | 50.5 | 55.3 |
| H3K4me2 | 25.8 | 25.8 | 31.3 | 36.5 | 31.1 | 51.2 |
| H3K4me3 | 20.5 | 23.1 | 28.9 | 40.3 | 36.3 | 58.5 |
| H3K79me3 | 46.3 | 54.1 | 60.1 | 64.7 | 67.4 | 73.0 |
| H3K9ac | 40.0 | 50.8 | 50.5 | 56.0 | 55.6 | 65.5 |
| H4 | 62.3 | 73.7 | 78.3 | 81.7 | 80.7 | 82.7 |
| H4ac | 25.5 | 38.4 | 38.6 | 49.1 | 50.4 | 61.7 |
| PD (avg.) | 77.1 | 35.0 | 84.6 | 88.1 | 84.2 | 82.3 |
| All | 75.8 | 47.4 | 90.4 | 91.0 | 86.8 | 86.0 |
| No-TATA | 85.1 | 52.2 | 93.6 | 94.0 | 94.3 | 93.7 |
| TATA | 70.3 | 5.3 | 69.8 | 79.4 | 71.6 | 67.4 |
| CPD (avg.) | 62.5 | 48.4 | 73.0 | 71.6 | 70.5 | 69.9 |
| All | 58.1 | 37.0 | 70.9 | 70.3 | 69.4 | 66.4 |
| No-TATA | 60.1 | 35.4 | 69.8 | 71.6 | 68.0 | 68.3 |
| TATA | 69.3 | 72.9 | 78.2 | 73.0 | 74.2 | 75.1 |
| SSD | 76.8 | 72.7 | 84.1 | 89.3 | 85.0 | 87.8 |
| COVID | 22.2 | 23.3 | 62.2 | 73.0 | 71.9 | 72.5 |
| Global Win % | 0.0 | 0.0 | 7.1 | 21.4 | 25.0 | 46.4 |
Safety Considerations
METAGENE-1 provides valuable capabilities for biosurveillance and genomic anomaly detection, showing state-of-the-art results on a broad coverage of benchmarks. While its current version poses minimal risk, we carefully weighed its benefits against potential misuse, particularly in synthetic biology, and emphasize the need for stricter safety considerations for future, more capable models.
Purpose and Capabilities: METAGENE-1 is specifically optimized to detect anomalies in short metagenomic reads (100-300 base pairs), making it well-suited for tasks like pathogen detection and biosurveillance. The model’s architectural constraints, such as its 512-token context length, limit its applicability to complex sequence design tasks, reducing misuse risks.
Open Source Impact: We believe the open release of METAGENE-1 will foster research in pathogen detection and biosurveillance by providing a valuable tool for scientists; it will also facilitate interpretability and controllability research in scientific foundation models. However, we emphasize the need for more rigorous safety evaluations before open-sourcing larger or more capable genomic models in the future.
We have included more in-depth discussions on safety considerations in our paper.
Model Details
- Release Date: Jan 06 2025
- Model License: Apache 2.0
BibTeX
@misc{liu2025metagene1metagenomicfoundationmodel,
title={METAGENE-1: Metagenomic Foundation Model for Pandemic Monitoring},
author={Ollie Liu and Sami Jaghouar and Johannes Hagemann and Shangshang Wang and Jason Wiemels and Jeff Kaufman and Willie Neiswanger},
year={2025},
eprint={2501.02045},
archivePrefix={arXiv},
primaryClass={q-bio.GN},
url={https://arxiv.org/abs/2501.02045},
}
- Downloads last month
- 21