|
--- |
|
tags: |
|
- monai |
|
- medical |
|
library_name: monai |
|
license: apache-2.0 |
|
--- |
|
# Model Overview |
|
Body CT segmentation models are evolving. Starting from abdominal multi-organ segmentation model [1]. Now the community is developing hundreds of target anatomies. In this bundle, we provide re-trained models for (3D) segmentation of 104 whole-body segments. |
|
|
|
This model is trained using the SegResNet [3] network. The model is trained using TotalSegmentator datasets [2]. |
|
|
|
 |
|
|
|
Figure source from the TotalSegmentator [2]. |
|
|
|
### MONAI Label Showcase |
|
|
|
- We highlight the use of this bundle to use and visualize in MONAI Label + 3D Slicer integration. |
|
|
|
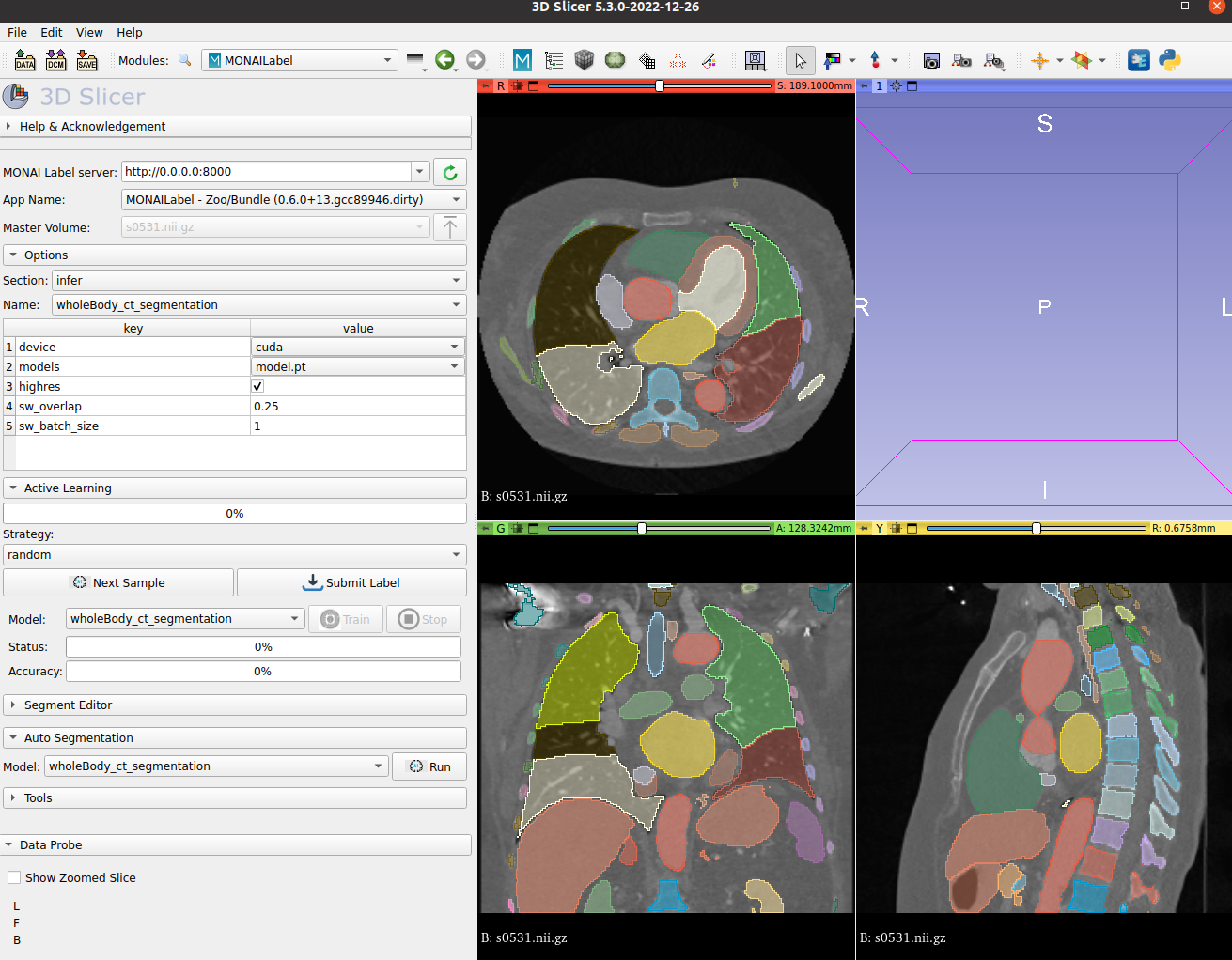 <br> |
|
|
|
## Data |
|
|
|
The training set is the 104 whole-body structures from the TotalSegmentator released datasets. Users can find more details on the datasets at https://github.com/wasserth/TotalSegmentator. All rights and licenses are reserved to the original authors. |
|
|
|
- Target: 104 structures |
|
- Modality: CT |
|
- Source: TotalSegmentator |
|
- Challenge: Large volumes of structures in CT images |
|
|
|
### Preprocessing |
|
|
|
To use the bundle, users need to download the data and merge all annotated labels into one NIFTI file. Each file contains 0-104 values, each value represents one anatomy class. We provide sample datasets and step-by-step instructions on how to get prepared: |
|
|
|
Instruction on how to start with the prepared sample dataset: |
|
|
|
1. Download the sample set with this [link](https://drive.google.com/file/d/1DtDmERVMjks1HooUhggOKAuDm0YIEunG/view?usp=share_link). |
|
2. Unzip the dataset into a workspace folder. |
|
3. There will be three sub-folders, each with several preprocessed CT volumes: |
|
- imagesTr: 20 samples of training scans and validation scans. |
|
- labelsTr: 20 samples of pre-processed label files. |
|
- imagesTs: 5 samples of sample testing scans. |
|
4. Usage: users can add `--dataset_dir <totalSegmentator_mergedLabel_samples>` to the bundle run command to specify the data path. |
|
|
|
Instruction on how to merge labels with the raw dataset: |
|
|
|
- There are 104 binary masks associated with each CT scan, each mask corresponds to anatomy. These pixel-level labels are class-exclusive, users can assign each anatomy a class number then merge to a single NIFTI file as the ground truth label file. The order of anatomies can be found [here](https://github.com/Project-MONAI/model-zoo/blob/dev/models/wholeBody_ct_segmentation/configs/metadata.json). |
|
|
|
## Training Configuration |
|
|
|
The segmentation of 104 tissues is formulated as voxel-wise multi-label segmentation. The model is optimized with the gradient descent method minimizing Dice + cross-entropy loss between the predicted mask and ground truth segmentation. |
|
|
|
The training was performed with the following: |
|
|
|
- GPU: 48 GB of GPU memory |
|
- Actual Model Input: 96 x 96 x 96 |
|
- AMP: True |
|
- Optimizer: AdamW |
|
- Learning Rate: 1e-4 |
|
- Loss: DiceCELoss |
|
|
|
## Evaluation Configuration |
|
|
|
The model predicts 105 channels output at the same time using softmax and argmax. It requires higher GPU memory when calculating |
|
metrics between predicted masked and ground truth. The consumption of hardware requirements, such as GPU memory is dependent on the input CT volume size. |
|
|
|
The recommended evaluation configuration and the metrics were acquired with the following hardware: |
|
|
|
- GPU: equal to or larger than 48 GB of GPU memory |
|
- Model: high resolution model pre-trained at a slice thickness of 1.5 mm. |
|
|
|
Note: there are two pre-trained models provided. The default is the high resolution model, evaluation pipeline at slice thickness of **1.5mm**, |
|
users can use the lower resolution model if out of memory (OOM) occurs, which the model is pre-trained with CT scans at a slice thickness of **3.0mm**. |
|
|
|
Users can also use the inference pipeline for predicted masks, we provide detailed GPU memory consumption in the following sections. |
|
|
|
### Memory Consumption |
|
|
|
- Dataset Manager: CacheDataset |
|
- Data Size: 1000 3D Volumes |
|
- Cache Rate: 0.4 |
|
- Single GPU - System RAM Usage: 83G |
|
- Multi GPU (8 GPUs) - System RAM Usage: 666G |
|
|
|
### Memory Consumption Warning |
|
|
|
If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate `cache_rate` in the configurations within range [0, 1] to minimize the System RAM requirements. |
|
|
|
### Input |
|
|
|
One channel |
|
- CT image |
|
|
|
### Output |
|
|
|
105 channels |
|
- Label 0: Background (everything else) |
|
- label 1-105: Foreground classes (104) |
|
|
|
## Resource Requirements and Latency Benchmarks |
|
|
|
### GPU Consumption Warning |
|
|
|
The model is trained with 104 classes in single instance, for predicting 104 structures, the GPU consumption can be large. |
|
|
|
For inference pipeline, please refer to the following section for benchmarking results. Normally, a CT scans with 300 slices will take about 27G memory, if your CT is larger, please prepare larger GPU memory or use CPU for inference. |
|
|
|
### High-Resolution and Low-Resolution Models |
|
|
|
We retrained two versions of the totalSegmentator models, following the original paper and implementation. |
|
To meet multiple demands according to computation resources and performance, we provide a 1.5 mm model and a 3.0 mm model, both models are trained with 104 foreground output channels. |
|
|
|
In this bundle, we configured a parameter called `highres`, users can set it to `true` when using 1.5 mm model, and set it to `false` to use the 3.0 mm model. The high-resolution model is named `model.pt` by default, the low-resolution model is named `model_lowres.pt`. |
|
|
|
In MONAI Label use case, users can set the parameter in 3D Slicer plugin to control which model to infer and train. |
|
|
|
- Pretrained Checkpoints |
|
- 1.5 mm model: [Download link](https://drive.google.com/file/d/1PHpFWboimEXmMSe2vBra6T8SaCMC2SHT/view?usp=share_link) |
|
- 3.0 mm model: [Download link](https://drive.google.com/file/d/1c3osYscnr6710ObqZZS8GkZJQlWlc7rt/view?usp=share_link) |
|
|
|
Latencies and memory performance of using the bundle with MONAI Label: |
|
|
|
Tested Image Dimension: **(512, 512, 397)**, the slice thickness is **1.5mm** in this case. After resample to **1.5** isotropic resolution, the dimension is **(287, 287, 397)** |
|
|
|
### 1.5 mm (highres) model (Single Model with 104 foreground classes) |
|
|
|
Benchmarking on GPU: Memory: **28.73G** |
|
|
|
- `++ Latencies => Total: 6.0277; Pre: 1.6228; Inferer: 4.1153; Invert: 0.0000; Post: 0.0897; Write: 0.1995` |
|
|
|
Benchmarking on CPU: Memory: **26G** |
|
|
|
- `++ Latencies => Total: 38.3108; Pre: 1.6643; Inferer: 30.3018; Invert: 0.0000; Post: 6.1656; Write: 0.1786` |
|
|
|
### 3.0 mm (lowres) model (single model with 104 foreground classes) |
|
|
|
GPU: Memory: **5.89G** |
|
|
|
- `++ Latencies => Total: 1.9993; Pre: 1.2363; Inferer: 0.5207; Invert: 0.0000; Post: 0.0358; Write: 0.2060` |
|
|
|
CPU: Memory: **2.3G** |
|
|
|
- `++ Latencies => Total: 6.6138; Pre: 1.3192; Inferer: 3.6746; Invert: 0.0000; Post: 1.4431; Write: 0.1760` |
|
|
|
## Performance |
|
|
|
### 1.5 mm Model Training |
|
|
|
#### Training Accuracy |
|
|
|
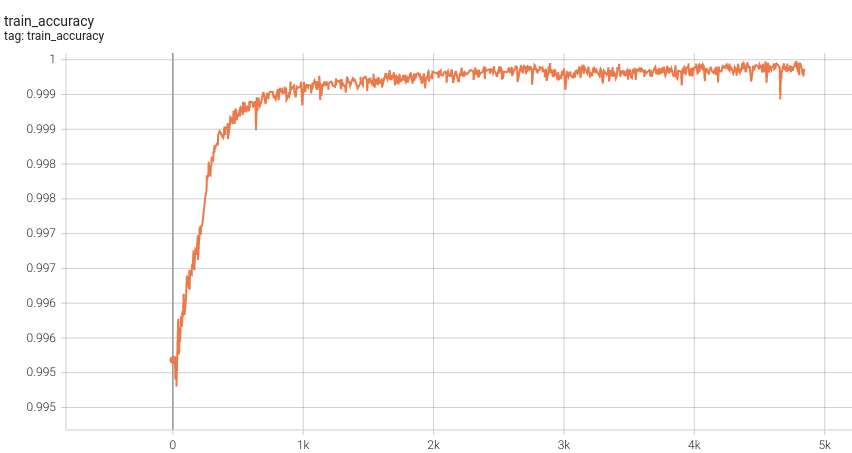 <br> |
|
|
|
#### Validation Dice |
|
|
|
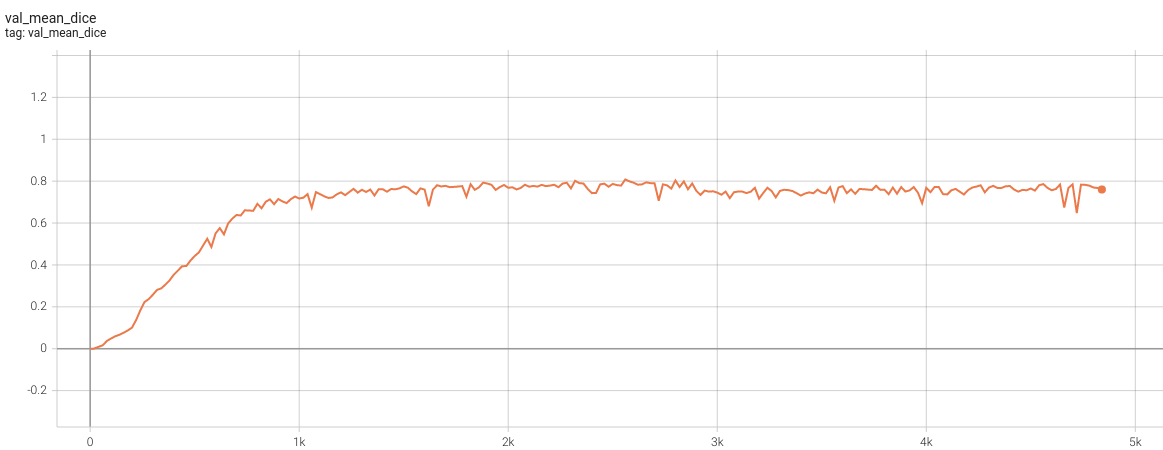 <br> |
|
|
|
Please note that this bundle is non-deterministic because of the trilinear interpolation used in the network. Therefore, reproducing the training process may not get exactly the same performance. |
|
Please refer to https://pytorch.org/docs/stable/notes/randomness.html#reproducibility for more details about reproducibility. |
|
|
|
## MONAI Bundle Commands |
|
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. |
|
|
|
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html). |
|
|
|
#### Execute training: |
|
|
|
``` |
|
python -m monai.bundle run --config_file configs/train.json |
|
``` |
|
|
|
Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using `--dataset_dir`: |
|
|
|
``` |
|
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path> |
|
``` |
|
|
|
#### Override the `train` config to execute multi-GPU training: |
|
|
|
``` |
|
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']" |
|
``` |
|
|
|
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove `--standalone`, modify `--nnodes`, or do some other necessary changes according to the machine used. For more details, please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html). |
|
|
|
#### Override the `train` config to execute evaluation with the trained model: |
|
|
|
``` |
|
python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']" |
|
``` |
|
|
|
#### Override the `train` config and `evaluate` config to execute multi-GPU evaluation: |
|
|
|
``` |
|
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json','configs/multi_gpu_evaluate.json']" |
|
``` |
|
|
|
#### Execute inference: |
|
|
|
``` |
|
python -m monai.bundle run --config_file configs/inference.json |
|
``` |
|
#### Execute inference with Data Samples: |
|
|
|
``` |
|
python -m monai.bundle run --config_file configs/inference.json --datalist "['sampledata/imagesTr/s0037.nii.gz','sampledata/imagesTr/s0038.nii.gz']" |
|
``` |
|
|
|
|
|
# References |
|
|
|
[1] Tang, Y., Gao, R., Lee, H.H., Han, S., Chen, Y., Gao, D., Nath, V., Bermudez, C., Savona, M.R., Abramson, R.G. and Bao, S., 2021. High-resolution 3D abdominal segmentation with random patch network fusion. Medical image analysis, 69, p.101894. |
|
|
|
[2] Wasserthal, J., Meyer, M., Breit, H.C., Cyriac, J., Yang, S. and Segeroth, M., 2022. TotalSegmentator: robust segmentation of 104 anatomical structures in CT images. arXiv preprint arXiv:2208.05868. |
|
|
|
[3] Myronenko, A., Siddiquee, M.M.R., Yang, D., He, Y. and Xu, D., 2022. Automated head and neck tumor segmentation from 3D PET/CT. arXiv preprint arXiv:2209.10809. |
|
|
|
|
|
|
|
# License |
|
|
|
Copyright (c) MONAI Consortium |
|
|
|
Licensed under the Apache License, Version 2.0 (the "License"); |
|
you may not use this file except in compliance with the License. |
|
You may obtain a copy of the License at |
|
|
|
http://www.apache.org/licenses/LICENSE-2.0 |
|
|
|
Unless required by applicable law or agreed to in writing, software |
|
distributed under the License is distributed on an "AS IS" BASIS, |
|
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. |
|
See the License for the specific language governing permissions and |
|
limitations under the License. |
|
|